One very simple application is the use of DNA markers to determine parentage. Although this might seem an obvious use of this technology, there are some important practical considerations to consider if you want to use DNA tests to sort out which bull sired which calf in a multi-sire breeding pasture.
The first is to make sure you collect a DNA sample (e.g., tail hair, blood card, semen) before the bulls are turned out with the cows. This might seem unimportant to do upfront, but it is sometimes hard to track down sires after the calves are born, and it is often the case that they are no longer available to provide a sample.
We recently evaluated bull performance in multi-sire breeding pastures using SNP-based DNA parentage testing for 5,052 calves from three ranches for three years and 15 calf crops in northern California.
Bulls averaged approximately 19 calves per calf crop, but that number varied greatly, ranging from zero to 64, with an average of 4.4 percent of the bulls in any breeding season siring no progeny. The genetics of those bulls siring a high number of calves were disproportionately represented in the early born replacement heifer pool.
Another application of DNA testing is to determine whether an animal is a carrier of a genetic condition. There are a number of recessive genetic conditions – meaning that an animal must inherit the same “a” allele from both parents (i.e., be homozygous) before there is an effect. Many genetic defects are recessive, and the reason for this is that mutant alleles often render the resulting protein non-functional.
In many cases, if an individual inherits a functional allele from one parent, there is no phenotype associated with inheriting the non-functional mutant allele from the other parent.
As such, a heterozygous “Aa” animal, or carrier, appears normal. (“A” symbolizes the dominant allele, “a” the recessive allele.) It is only when two carriers mate there is the possibility of producing offspring that have by chance inherited both of the non-functional alleles from their parents.
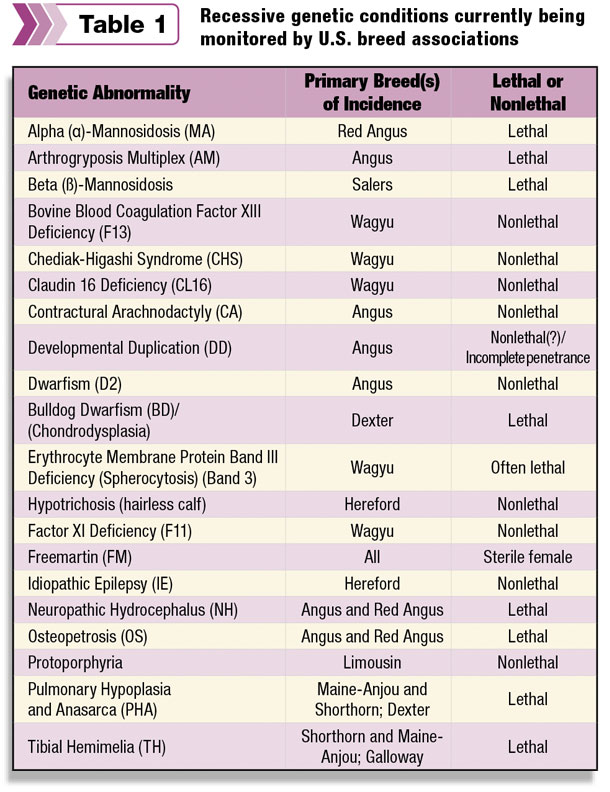
Table 1 lists the recessive genetic conditions currently being monitored by U.S. beef breed associations.
There are many management options for the control of genetic conditions, and the best choice will depend on the needs and requirements of each enterprise. Some of these options include:
1. Testing all animals and culling carriers
2. Testing all animals and using carriers only in terminal breeding programs
3. Testing sires and using only free bulls for breeding
This last option will eliminate affected progeny and decrease the number of carriers over time. However, in some cases, the overall breeding value of carrier animals may outweigh the economic penalty associated with their carrier status.
Carrier sires and dams can be used for breeding. However, follow-up testing of all progeny would be essential to develop future breeding strategies with that progeny because we would expect 50 percent of their progeny to be carriers.
These carriers can be identified by DNA testing. They may be culled or used in a separate breeding program with animals of identified negative or free status. The negative-status progeny can then be used to perpetuate phenotypically and quantitatively selected superior production traits. Continued breeding by mating two carriers risks the production of approximately 25 percent affected animals.
Another use of DNA information is to improve the accuracy of genetic merit estimates, typically EPDs. It has become clear genomic tests for this purpose are breed-specific, meaning that if a genomic test is developed using data from one breed, it will not be accurate at predicting traits in a different breed.
Consequently, a number of breed associations have developed their own breed-specific training populations (a database containing thousands of genotyped animals from that breed and their associated performance records on traits of interest) from which to develop genomic predictions that can be used to improve the accuracy of EPD within that breed.
These “genomic-enhanced EPDs” (GE-EPDs) include the DNA markers as an additional source of information, which improves the accuracy of the EPDs, especially in young unproven animals where the accuracy is typically only provided by a pedigree estimate (i.e., interim EPDs) and possibly the bull’s own record. The beef breed associations that currently offer GE-EPDs include Angus, Gelbvieh, Hereford, Simmental, Limousin and Red Angus.
According to Dr. Dorian Garrick, Lush Chair in Animal Breeding and Genetics at Iowa State University and executive director of the National Beef Cattle Evaluation Consortium (NBCEC), other beef breed associations that are working to develop GE-EPDs include Charolais, Shorthorn, Maine-Anjou, Santa Gertrudis and Brangus.
Determining which animals to test is a question of economics. The cost of the high-density test for GE-EPDs is in the vicinity of $75 to $80 per test. The value derived from the test is improved accuracy of EPDs in young animals.
Several large USDA-funded projects are now targeting the development of DNA tests that will help provide selection criteria for hard-to-measure traits such as feed efficiency, disease resistance and fertility. It is hoped these projects will result in products that will enable these economically important traits to be included in beef cattle selection decisions in the future. ![]()

- Alison Van Eenennaam
- Genomics
- University of California – Davis








